Plasmid-mediated quinolone resistance determinants in clinical bacterial pathogens isolated from the Western Region of Ghana: a cross-sectional study
Andrews Kwabena Sah, Patrick Kwame Feglo
Corresponding author: Patrick Kwame Feglo, Department of Clinical Microbiology, School of Medicine and Dentistry, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana 
Received: 02 Mar 2022 - Accepted: 14 Dec 2022 - Published: 27 Dec 2022
Domain: Bacteriology,Microbiology,Infectious diseases epidemiology
Keywords: Antimicrobial resistance, quinolones, plasmid-mediated quinolone resistance genes, Ghana
©Andrews Kwabena Sah et al. Pan African Medical Journal (ISSN: 1937-8688). This is an Open Access article distributed under the terms of the Creative Commons Attribution International 4.0 License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article: Andrews Kwabena Sah et al. Plasmid-mediated quinolone resistance determinants in clinical bacterial pathogens isolated from the Western Region of Ghana: a cross-sectional study. Pan African Medical Journal. 2022;43:207. [doi: 10.11604/pamj.2022.43.207.33734]
Available online at: https://www.panafrican-med-journal.com//content/article/43/207/full
Research 
Plasmid-mediated quinolone resistance determinants in clinical bacterial pathogens isolated from the Western Region of Ghana: a cross-sectional study
Plasmid-mediated quinolone resistance determinants in clinical bacterial pathogens isolated from the Western Region of Ghana: a cross-sectional study
&Corresponding author
Introduction: quinolones are critically important antibiotics that are reserved for treating very severe infections caused by multidrug-resistant bacterial pathogens. However, their indiscriminate uses have resulted in an increased number of resistant strains in many parts of the world including Ghana. We determined the quinolone resistance profile of Gram-negative bacterial pathogens and characterized the underlying molecular determinants of resistance.
Methods: Gram-negative pathogens obtained from clinical specimens at three hospital laboratories were tested for resistance to quinolones and other commonly used antibiotics. ESBL production among the Enterobacteriale isolates was confirmed using the combined disc diffusion method. We then used PCR to determine seven types of plasmid-mediated quinolone resistance genes present in the isolates resistant to nalidixic acid and ciprofloxacin.
Results: in this study, 29.5% of the isolates were resistant to ciprofloxacin, with the highest of 50% among E. coli resistance to the other quinolones was levofloxacin (24.4%), norfloxacin (24.9%), and nalidixic acid (38.9%). Significant proportions of the quinolone-resistant isolates were ESBL producers (P-values < 0.001). The aac(6´)-Ib-cr, qnrS, oqxA, and qepA genes were present in 43 (89.6%), 27 (56.3%), 23 (47.9%), and one (2.1%) of the isolates, respectively. None of the isolates tested positive to qnrA, qnrB, and oqxB genes. The presence of the aac(6´)-Ib-cr gene positively correlated with resistance to ceftriaxone, cefotaxime, and gentamicin (P-values <0.05).
Conclusion: high proportions of Gram-negative bacterial isolates were resistant to quinolones and most of these isolates possessed multiple PMQR genes. There is a need to implement measures to limit the spread of these organisms.
Antibiotics are essential drugs for controlling and preventing morbidity and mortality from infection caused by bacteria. However, rising resistance levels in nearly all bacteria types that cause illnesses have substantially reduced their effectiveness and threatened a return to the pre-antibiotic era [1]. This has led to increased length of hospital stays, cost of healthcare, morbidity and death [2]. Quinolones are an important group of these essential drugs and were introduced into clinical practices in the mid-1980s. They are well known for their ease of absorption following oral administration, with higher concentrations in the urinary tract. This often makes them the drug of choice for urinary tract infections. Newer generations such as ciprofloxacin are mostly preferred in treating infections such as enteric fevers to other groups which are required to be administered parenterally, including the cephalosporins and aminoglycosides [3]. Quinolones are also one of the major options for the treatment of infections caused by dangerous and multidrug-resistant bacteria [1]. In Ghana, quinolones were introduced for the treatment of bacterial infections due to increased resistance levels in bacterial pathogens to commonly used, less expensive, and readily available antibiotics such as the penicillins, sulfonamides and first and second-generation cephalosporins [1,4]. However, the recurrence of bacterial resistance to this family of antibiotics is limiting their use globally, whiles alternatives are limited. Urgent measures are therefore required to reduce the spread of resistant bacteria. This will help reduce the already existing pressure on health systems in resource-limited countries and promote the achievement of the Sustainable Development Goals (SDGs) [2].
Resistance to the quinolones is generally higher in developing countries. A systematic review in 2015 recorded levels as low as 2% in USA and Ireland, compared to 80% in Nigeria [5]. In Ghana, up to 50% resistance to ciprofloxacin was reported among pathogens obtained from wound and blood cultures [6,7]. In particular, there has been increasing reports of reduced susceptibility of Salmonella species to fluoroquinolones [4,8,9]. Resistance to quinolone antibiotics is mediated by chromosomal mutations and the acquisition of transferable plasmid genes [3,10,11]. Several forms of plasmid-mediated quinolone resistance (PMQR) genes have been described and most of these have been detected in clinical isolates [3]. These genes facilitate the selection and spread of quinolone-resistant bacterial strains rapidly and pose a greater threat to public health [10,12]. Studies in most countries have determined the prevalence and characteristics of the various resistant determinants in clinical isolates. In Ghana, there is a dearth of studies on quinolone resistance determinants in clinical isolates and the few studies are centered and localized to teaching hospital in the two big cities in the country; Accra and Kumasi. This study aimed to determine the molecular determinants that mediate quinolone resistance among Gram-negative bacterial pathogens in the Western Region of Ghana. Data obtained from the study are essential to inform policy on antibiotic stewardship in the region and to monitor the effectiveness of existing interventions.
Study design, sites, and populations: a prospective cross-sectional study was conducted in the Western Region of Ghana from October 2020 to February 2021. The region is situated in the Southwestern part of Ghana. It shares borders with the Central Region in the East, Western North Region in the North, La Cote d´Ivoire in the West, and the Gulf of Guinea (Atlantic Ocean) in the South [13]. The Region is divided into fourteen administrative divisions, including one Metropolitan, seven Municipalities, and six Districts [14]. Clinical specimens of patients conducting microbiology tests at the Sekondi Public Health Laboratory (SPHL), situated in the Western Regional capital; Tarkwa Municipal Hospital (TMH), in a semi-urban city; and Prestea Government Hospital (PGH), in a rural town were obtained for the study. Gram-negative bacterial pathogens isolated from Mid-stream urine, blood and wound swab specimens were included in the study.
Bacterial isolation and identification: the specimen were cultured using conventional techniques [15]. We used cysteine-lactose-electrolyte-deficient (CLED) agar for culturing the urine specimens. Blood and wound specimens were cultured on MacConkey agar and blood agar. The inoculated plates were incubated aerobically at 35-37°C and examined after 18-24 hours for bacterial growth. For the urine cultures, a count of 105CFU/mL or more bacterial growth was considered significant for further investigations. The bacterial pathogens were identified using conventional methods including their Gram staining reactions, colonial characteristics, and biochemical properties.
Antibiotic susceptibility testing: Gram-negative isolates were selected for antimicrobial susceptibility testing using the modified Kirby-Bauer disc diffusion method in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines [16]. The antimicrobials tested included ciprofloxacin (5µg), levofloxacin (5µg), amikacin (30µg), piperacillin-tazobactam (110µg), gentamicin (10µg) ceftazidime (30µg), meropenem (10µg). nalidixic acid (30µg), norfloxacin (10µg), ampicillin-sulbactam (20µg), amoxicillin-clavulanic acid (30µg), cefotaxime (30µg), nitrofurantoin (300µg), ceftriaxone (30µg), and trimethoprim-sulfamethoxazole (25µg). Among the enterobacteriaceae, isolates that tested resistant to cefotaxime, ceftazidime, or ceftriaxone were suspected to be ESBL producers [16], and were confirmed using the combination disc method [16].
Detection of plasmid-mediated quinolone resistance genes using polymerase chain reaction (PCR): following the manufacturer´s instructions, we extracted plasmid DNA from 48 isolates resistant to ciprofloxacin and nalidixic acid using ZymoPURE™ Plasmid Miniprep kit (Zymo Research Corporation). We used qualitative conventional polymerase chain reaction (PCR) carried out on TC-512 (Techne, UK) thermocycler to amplify seven plasmid-mediated quinolone resistance genes. The genes and the primers used were qnrA (forward 5´- AGAGGATTTCTCACGCCAGG-3´ and reverse 5´- GCAGCACTATKACTCCCAAGG-3´), qnrB (forward 5´ GGMATHGAAATTCGCCACTG-3´ and reverse 5´- TTTGCYGYYCGCCAGTCGAA-3´), qnrS (forward 5´- GCAAGTTCATTGAACAGGCT-3´ and reverse 5´- TCTAAACCGTCGAGTTCGGCG-3´), qepA (forward 5´- CTGCAGGTACTGCGTCATG-3´ and reverse 5´- CGTGTTGCTGGAGTTCTTC-3´), OxqA (forward 5´-GACAGCGTCGCACAGAATG-3´ and reverse 5´-GGAGACGAGGTTGGTATGGA-3´), oxqB (forward 5´-CGAAGAAAGACCTCCCTACCC-3´ and reverse 5´-CGCCGCCAATGAGATACA-3´), and aac(6´)-Ib-cr (forward 5´-TTGCGATGCTCTATGAGTGGCTA-3´ and reverse 5´-CTCGAATGCCTGGCGTGTTT-3´) [17]. A 25 µl reaction volume consisting of 9µl nuclease-free water, 0.5 µl of 10 µm forward primer, 0.5 µl of 10 µm reverse primer, 12.5 µl of ‘One-Taq Quick Load 2X Mater Mix with Standard Buffer´ and 2.5 µl of template DNA was set up. The PCR was run using the following cycling conditions: initial denaturation at 94°C for 30 seconds; 30 cycles of denaturation at 94°C for 30 seconds, annealing for 30 seconds at 55°C for qnrA, qnrB, qnrS and aac(6´)-1b-cr and at 54°C for qepA, oqxA and oqxB, and extension at 68°C for 60 seconds; final extension at 68°C for 5 minutes; and holding at 4°C till removed from the thermocycler. The amplicons were resolved using electrophoresis on 1.5% agarose gel at 5V/cm for 60 minutes.
Data analysis: the data generated from the study were captured into Microsoft Excel 2013 spreadsheet and exported into IBM SPSS Statistics 23 for analysis. The antibiotic sensitivity results (zone diameters) were captured, analyzed and interpreted using the 2020 version of WHONET software. The results were summarized into tables and charts. Categorical variables were presented as frequencies and proportions (%), and continuous variables as mean and median. A chi-square test was used to compare the categorical variables while a student T-Test was used for continuous variables. A p-value of ≤0.05 was considered significant.
Ethical considerations: the Committee on Human Research Publication and Ethics of the School of Medicine and Dentistry at Kwame Nkrumah University of Science and Technology approved the study protocol with reference number CHRPE/AP/302/20. The authorities of all the participating facilities also granted permission for sample collection and access to laboratory units. Informed consent was obtained from the participants by the health professional collecting the samples after adequate information about the project was explained to them in accordance with the Declaration of Helsinki.
Demographic data: between October 2020 and February 2021, 820 specimens were collected from 807 patients (591 females and 216 males). The specimens included urine 538 (65.6%), blood 180 (22%), and wound 102 (12.4%). Most of the specimens 376 (44.8%) were collected at Sekondi Public Health Laboratory, 203 (24.8%) at Tarkwa Municipal Hospital, and 237 (28.9%) at Prestea Government Hospital (Table 1). The ages of the participants ranged from 2 days to 94 years, with a median age of 28. Most of the participants, 572 (70.1%), sought outpatient services, whereas in-patients constituted 235 (19.1%).
Bacteria isolated: among the 807 participants, the samples from 258 were culture-positive for Gram-negative bacteria, giving a prevalence of 32.0%. These 258 specimens yielded 272 bacterial isolates in total, out of which 254 were used for further analysis (Table 1). Majority of the isolates were Klebsiella spp. 71 (26.1%) and E. coli 70 (25.7%).
Quinolone antimicrobial resistance pattern of the isolates: antimicrobial susceptibility testing was performed on 254 isolates. The proportions of the isolates that were resistant to the four tested quinolones in descending order were nalidixic acid 86/221 (38.9%), ciprofloxacin 75/254 (29.5%), norfloxacin 55/221 (24.9%), and levofloxacin 62/254 (24.4%). Among the isolates, the highest proportions of resistance to the quinolones were recorded in E. coli followed by Klebsiella spp., while Salmonella spp. and Proteus spp. had the least resistance proportions (Table 2). The proportions of E. coli and Klebsiella spp. resistant to the tested quinolones were compared; the results indicated that E. coli isolates had higher resistance proportions than Klebsiella spp. and the differences were statistically significant (p-values < 0.05). Among patient groups and sexes there were no significant differences in resistance proportions to any of the quinolones tested (P-values > 0.05). In this study, geographical differences in quinolone resistance were observed. Although, isolates from Sekondi Public Health Laboratory and Tarkwa Municipal Hospital showed no significant differences in resistance to the tested quinolones, they had significantly higher levels of resistance to all the four quinolones than isolates from Prestea Government Hospital (P values < 0.05) (data not shown).
Resistance patterns of the isolates to other antimicrobials: the isolates were tested against eleven other antimicrobials. The highest proportion of the isolates were resistant to trimethoprim sulfamethoxazole 143 (64.7%), followed by amoxicillin clavulanic acid 103 (46.6%), ceftriaxone 95 (42.5%), and cefotaxime 94 (42.5%), the least proportions of resistance were to amikacin 5 (2.0%) and meropenem 12 (4.7%) (Table 2).
Extended spectrum beta-lactamase (ESBL) production as a predictor of resistance to quinolones : we tested 215 Enterobacteriaceae isolates for ESBL production and 77 (35.8%) were confirmed as ESBL producers. The highest proportion of ESBL producers was observed among E. coli 34/70 (48.6%), followed by Klebsiella spp. 27/69 (39.1%), and Citrobacter spp. 6/19 (31.6%). None of the Proteus spp. was ESBL producer. There was a link between quinolone-resistance and ESBL production. The proportions of ESBL producing isolates and the non-ESBL producing isolates resistant to the tested quinolones were compared. Isolates that produce ESBL were more likely to be resistant to the quinolone antibiotics than their non-ESBL producing counterparts (odd ratios > 43, P-values < 0.0001).
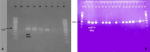
Plasmid-mediated quinolone resistant genes : we screened forty-eight (48) samples, made up of 27 strains of E. coli, 16 strains of Klebsiella spp., four of Citrobacter spp., and one Enterobacter sp. for PMQR genes using conventional PCR. Out of this, 46 (95.8%) tested positive for at least one of the PMQR genes, 30 (62.5%) samples contained multiple genes with 15 (31.3%) harboring three (3) genes. The plasmid genes detected were aac(6´)-Ib-cr 43 (89.6%), qnrS 27 (56.3%), oqxA 23 (47.9%), and qepA one (2.1%). We did not detect the qnrA, qnrB, and oqxB genes in any of the samples tested. Figure 1 shows examples of gel images of PCR products captured. Among the 27 strains of E. coli. tested, aac(6´)-Ib-cr, qnrS, oqxA and qepA were present in 22 (81.5%), 19 (70.4%), 17 (63.0%), andone (3.7%) respectively. All the Klebsiella spp., Citrobacter spp., and Enterobacter sp. tested possessed aac(6´)-Ib-cr gene. One (25%) of Citrobacter species possessed qnrS and oqxA genes. Among the Klebsiella strains, six (37.5%) and five (3.3%) possessed qnrS and oqxA genes respectively while among Citrobacter spp. qnrS, one (25%) and oqxA, one (25%) genes were present. The Enterobacter sp. also possessed qnrS gene.
The presence of PMQR genes was investigated to determine whether it affects susceptibility to antibiotics other than quinolones (Table 3). We observed that the presence of qnrS and oqxA genes did not correlate with the proportion of the isolates that were resistant to any of the antimicrobials tested (p-values > 0.05). However, the presence of aac(6´)-Ib-cr gene positively correlated to the proportions of the isolates that were resistant to cefotaxime (P-value=0.006), ceftriaxone (p-value=0.018), and gentamicin (p-value=0.020).
Antimicrobial resistance has emerged as an important threat to human and animal health worldwide, calling for intensive research to elucidate the complexity of underlying causes and possible ways of reducing, stopping, and possibly reversing the rate of its development [18,19]. In this study, we studied the antibiotic resistance patterns of 254 clinical Gram-negative bacterial isolates collected from three hospitals in the Western Region of Ghana. The presence of plasmid-mediated quinolone resistance genes was determined in selected ciprofloxacin and nalidixic acid resistant isolates. The overall prevalence of Gram-negative bacterial pathogens from all the clinical specimens was 32.0% in the current study. This is similar to the finding of a study from Ethiopia where 34.7% of specimens had Gram-negative bacteria recovered from them [20]. However, it is higher when compared to results of studies from Mexico and Nepal, where prevalence of 19.1% and 17%, respectively, were reported [21,22]. These observed differences could be attributed to the differences in study settings, choice of study populations, sources of samples, and sample sizes. Among the thirteen different Gram-negative bacterial species identified, E. coli and Klebsiella species were the most dominant. They constituted 51.8% of the total isolates, as shown in Table 1. This is similar to the results of several studies, showing that, among Gram-negative bacterial pathogens, E. coli and Klebsiella spp. are frequent cause of infections in humans. These pathogens are becoming difficult to treat due to the development of resistance to the readily available and less costly drugs used by health professionals to treat them [20,22-24].
In this study, resistance patterns of thirteen bacterial species to fifteen antibiotics belonging to six different antibiotic classes were studied. Generally, high proportions of the isolates were resistant to the commonly prescribed antibiotics except for amikacin and meropenem where resistance proportions were below 5%. This result is similar to the findings of Agyepong and colleagues who found resistance proportions of Gram-negative bacteria isolated from a teaching hospital in Ghana at 3.5% and 2.5% to amikacin and meropenem, respectively [23]. Meropenem and Amikacin are relatively expensive making them not readily available, limiting their use in the study population. In addition, these drugs are recently introduced into the Ghanaian market and are mostly used as a last resort for treating multidrug-resistant infections. This has possibly led to relatively low natural selection and hence low development of resistance among the isolates. However, the emergence of resistance to these drugs especially, the carbapenems is worrying [25] and if unchecked can become a greater public health challenge in the future. Quinolone antibiotics are widely used to treat various forms of bacterial infections since being introduced in Ghana in the past two decades [4]. Due to their lower prices, accessibility, and ease of administration, these drugs have extensively been abused [26]. In a recent multicenter point prevalence survey across seven hospitals in Ghana, ciprofloxacin was among the top five most prescribed antibiotics to in-patients. The survey found 10% of these prescriptions to have no documented reasons [27]. The increased and inappropriate use of these drugs [28] has resulted in an increased selection for resistant bacteria strains. In this study, overall proportion of isolates that were resistant to ciprofloxacin was found to be 29.5% ranging from 0% in Proteus spp. and Salmonella spp. to 50% in E. coli. This is similar to the results of other studies where resistance of Gram-negative bacterial pathogens to ciprofloxacin were found to be 25% [29] and 26.7% [30]. Although, the proportions of resistance to ciprofloxacin were low compared to other studies [20,23,31], it is relatively high compared to findings from other countries, especially those in Europe and North America [32,33]. These observed differences were probably due to differences in geographical distribution of resistant bacterial strains [34], study population, study design [35], existing drug prescription policies, AMR surveillance, and antimicrobial stewardship programs [26,36]. Infection with a quinolone-resistant and/or ESBL producing organism has significant public health implications. These infections have limited treatment options, making it difficult for clinicians to prescribe successful treatment. They also make empiric prescriptions perilous since they are often associated with poorer treatment outcomes, including increased treatment failure, mortality, and morbidity [26,37]. They also increase the pressure on health facilities, which are often inadequate in our settings, due to increased hospital stays and the need for additional procedures due to complications [38]. Invariably, these translate into a higher social and economic burden on individuals, families, and the community.
In the present study, four PMQR genes were detected in decreasing order: aac(6´)-Ib-cr 43 (89.6%), qnrS 27 (56.2%), oqxA 23 (47.9%), and qepA one (2.1%) among ciprofloxacin and nalidixic acid resistant isolates. The predominance of the aac(6´)-Ib-cr gene is supported by the work of Attipoe et al. involving E. coli isolated from 18 testing laboratories across Ghana. In their work, all 29 isolates tested were positive for the aac(6´)-Ib-cr gene. They also found a larger proportion of E. coli isolates to possess qnrS 26 (89.6%), oqxA 19 (65.5%), qnrA 16 (55.1%), and qnrB 15 (55.1%) [39]. Contrarily, the present study did not detect qnrA, qnrB and oqxB in the isolates. This may be due to differences in the geographical distribution of PMQR genes. Other studies in Nigeria [40] and Iran [41] confirmed the predominance of the aac(6´)-Ib-cr gene. These reports indicate the widespread presence of this quinolone-resistance gene.
In this study, no correlation was observed between the presence of qnrS and oqxA genes and the level of resistance to the non-quinolone antibiotics tested. This implies that, the presence of these genes alone may not be enough to affect the efficacy of the non-quinolone antibiotics. Isolates possessing aac(6´)-Ib-cr genes were found to have significant levels of resistance to cefotaxime (P-value = 0.006), ceftriaxone (P-value = 0.018), and gentamicin (P-value = 0.020) as compared to their aac(6´)-Ib-cr negative counterparts. No correlation was found with the other antibiotics. The enzyme aminoglycoside acetyltransferase, encoded by the aac(6´)-Ib-cr gene is originally known to confer resistance to the aminoglycosides (tobromycin, kanamycin, amikacin and gentamicin). Hence resistance to gentamycin in the presence of aac(6´)-Ib-cr gene is expected. The aac(6´)-Ib-cr gene is found in various integrons, especially on IncF11 plasmids that express CTX-M-15 [12]. CTX-M-15 is a widespread ESBL that confers resistance to third generation cephalosporin such as ceftriaxone and cefotaxime. Hence, isolates with aac(6´)-Ib-cr genes are likely to be resistance to third generation cephalosporin. These may explain the observed association between the presence of aac(6´)-Ib-cr gene and resistance to ceftriaxone and cefotaxime [12].
Limitations of this study: sequencing of PMQR genes identified in the study was not done to determine the allelic forms present in the study areas. Minimum inhibition concentration (MIC) determination for quinolone antibiotics was not done. MIC could have helped to determine if there is difference in levels of resistance to quinolone among the isolates positive and negative for PMQR genes. In addition, this is a laboratory based study so risk factors such as previous admission to hospital, length of stay at the hospital, presence of indwelling devices, and history of antibiotic use could not be assessed.
In this study, high proportion of quinolone-resistant Gram-negative bacterial pathogens possessed multiple horizontally transferable PMQR genes. These genes promote the selection of highly quinolone-resistant bacterial strains that when spread can pose serious threat to public health in the region and Ghana. It is therefore recommended to the regional health authorities and stakeholders to institute measures to ensure adequate antimicrobial stewardship and surveillance, adequate infection prevention and control, and good prescription behavior among health workers to prevent further spread of quinolone resistant bacterial pathogens.
What is known about this topic
- The role of plasmid-mediated quinolone resistance genes in the development of bacterial resistance to quinolone antibiotics;
- The circulation of plasmid-mediated quinolone genes in the gram-negative bacterial population of Ghana;
- The production of extended-spectrum beta-lactamase as predictor of quinolone antimicrobial resistance in gram-negative bacterial.
What this study adds
- The prevalence of aac(6´)-Ib-cr, qnrS, oqxA, and qepA among quinolone-resistant gram-negative bacterial pathogens isolated from the Western Region of Ghana;
- The prevalence of quinolone antimicrobial resistance among gram-negative bacterial pathogens isolated from the Western Region of Ghana.
The authors declare no competing interests.
Andrews Kwabena Sah carried out the work and wrote the manuscript. Patrick Kwame Feglo developed the research concept, supervised the work, and edited the manuscript. All the authors have read and agreed to the final version of the manuscript.
The authors appreciate the staff of Sekondi Public Health Laboratory, Tarkwa Municipal Hospital Laboratory, and Prestea Government Hospital Laboratory for their technical support and guidance during the fieldwork. More especially, to Mr Ebenezer Kofi Mensah, Mr. Solomon Asante-Safa, Mr Emixon Essuman, Mr Frank Angate and Dr. Emmanuel D. Darko. We also extend our profound gratitude to Dr P. Narkwah of the Virology Laboratory of Department of Clinical Microbiology, KNUST for his support during the molecular workout at his laboratory.
Table 1: the distribution of bacterial pathogens isolated from the Western Region of Ghana, stratified by gender, patient category, age, specimen type, and testing laboratory
Table 2: antibiotic resistance pattern of Gram-negative pathogens isolated from patients at three laboratories in the Western Region of Ghana
Table 3: plasmid-mediated quinolone-resistant genes as predictors of resistance to non-quinolone antibiotics
Figure 1: agarose gel electrophoresis of PCR products; A-oqxA; B-aac(6´)-Ib-cr
- Collignon PC, Conly JM, Andremont A, McEwen SA, Aidara-Kane A. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin Infect Dis. 2016;63(8):1087-1093. PubMed | Google Scholar
- Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet. 2022;399(10325):629-655. PubMed | Google Scholar
- Kim ES, Hooper DC. Clinical importance and epidemiology of quinolone resistance. Infect Chemother. 2014;46(4):226. PubMed | Google Scholar
- Eibach D, Al-Emran HM, Dekker DM, Krumkamp R, Adu-Sarkodie Y, Cruz Espinoza LM et al. The emergence of reduced ciprofloxacin susceptibility in Salmonella enterica causing bloodstream infections in rural Ghana. Clin Infect Dis. 2016;62(suppl 1):S32-S36. PubMed | Google Scholar
- Fasugba O, Gardner A, Mitchell BG, Mnatzaganian G. Ciprofloxacin resistance in community- and hospital-acquired Escherichia coli urinary tract infections: a systematic review and meta-analysis of observational studies. BMC Infect Dis. 2015;15:545. PubMed | Google Scholar
- Janssen H, Janssen I, Cooper P, Kainyah C, Pellio T, Quintel M et al. Antimicrobial-resistant bacteria in infected wounds, Ghana, 20141. Emerg Infect Dis. 2018;24(5):916-919. PubMed | Google Scholar
- Groß U, Amuzu SK, de Ciman R, Kassimova I, Groß L, Rabsch W et al. Bacteremia and antimicrobial drug resistance over time, Ghana. Emerg Infect Dis. 2011;17(10):1879-1882. PubMed | Google Scholar
- Acheampong G, Owusu M, Owusu-Ofori A, Osei I, Sarpong N, Sylverken A et al. Chromosomal and plasmid-mediated fluoroquinolone resistance in human Salmonella enterica infection in Ghana. BMC Infect Dis. 2019;19(1):898. PubMed | Google Scholar
- Andoh LA, Ahmed S, Olsen JE, Obiri-Danso K, Newman MJ, Opintan JA et al. Prevalence and characterization of Salmonella among humans in Ghana. Trop Med Health. 2017;45(1):3. PubMed | Google Scholar
- Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. The Lancet. 1998;351(9105):797-799. PubMed | Google Scholar
- Correia S, Poeta P, Hébraud M, Capelo JL, Igrejas G. Mechanisms of quinolone action and resistance: where do we stand? J Med Microb. 2017;66(5):551-559. PubMed | Google Scholar
- Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22(4):664-689. PubMed | Google Scholar
- Ghana Statistical Services. Ghana Statistical Service. Ghana. 2021. Accessed 11 September 2021
- Western Region, Community Water and Sanitation Agency. Regional profile: population, location and size. Community Water and Sanitation Agency. 2020. Accessed 11 April 2022
- Vandepitte J, Verhaegen J, Engbaek K, Rohner P, Piot P, Heuck CC (eds). Basic laboratory procedures in clinical bacteriology. Geneva, World Health Organization. 2nd ed: 2003. Google Scholar
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing: A CLSI supplement M100. Clinical and Laboratory Standard Institute. 29th ed: 2020.
- Chen X, Zhang W, Pan W, Yin J, Pan Z, Gao S et al. Prevalence of qnr, aac(6 ´ )-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother. 2012;56(6):3423-3427. PubMed | Google Scholar
- World Health Organization. Global antimicrobial resistance and use surveillance system (GLASS) report: 2021. Geneva, World Health Organization. 2021. Google Scholar
- The Lancet. The antimicrobial crisis: enough advocacy, more action. The Lancet. 2020;395(10220):247. PubMed | Google Scholar
- Moges F, Eshetie S, Abebe W, Mekonnen F, Dagnew M, Endale A et al. High prevalence of extended-spectrum beta-lactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS ONE. 2019;14(4):1-13. PubMed | Google Scholar
- Ghimire A, Acharya B, Tuladhar R. Extended spectrum ß-lactamase (ESBL) producing multidrug resistant Gram-negative bacteria from various clinical specimens of patients visiting a tertiary care hospital. TU J Microbiol. 2018;4:1-8. Google Scholar
- Uc-Cachón AH, Gracida-Osorno C, Luna-Chi IG, Jiménez-Guillermo JG, Molina-Salinas GM. High prevalence of antimicrobial resistance among Gram-negative isolated bacilli in intensive care units at a tertiary-care hospital in Yucatán Mexico. Medicina. 2019;55(9):588. PubMed | Google Scholar
- Agyepong N, Govinden U, Owusu-Ofori A, Essack SY. Multidrug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob Resist Infect Control. 2018;7:37. PubMed | Google Scholar
- Feglo PK, Adu-Sarkodie Y. Antimicrobial resistance patterns of extended spectrum ß-lactamase producing Klebsiellae and E. coli isolates from a tertiary hospital in Ghana. ESJ. 2016;12(30):174. PubMed | Google Scholar
- Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Therapeutic Advances in Infection. 2016;3(1):15-21. PubMed | Google Scholar
- Iskandar K, Molinier L, Hallit S, Sartelli M, Catena F, Coccolini F et al. Drivers of antibiotic resistance transmission in low- and middle-income countries from a “One Health” perspective:a review. Antibiotics. 2020;9(7):372. PubMed | Google Scholar
- Labi A-K, Obeng-Nkrumah N, Dayie NTKD, Egyir B, Sampane-Donkor E, Newman MJ et al. Antimicrobial use in hospitalized patients: a multicentre point prevalence survey across seven hospitals in Ghana. JAC-Antimicrobial Resistance. 2021;3(3):dlab087. PubMed | Google Scholar
- Bediako-Bowan AAA, Owusu E, Labi A-K, Obeng-Nkrumah N, Sunkwa-Mills G, Bjerrum S et al. Antibiotic use in surgical units of selected hospitals in Ghana: a multi-centre point prevalence survey. BMC Public Health. 2019;19(1):797. PubMed | Google Scholar
- Karikari AB, Saba CKS, Yamik DY. Assessment of asymptomatic bacteriuria and sterile pyuria among antenatal attendants in hospitals in northern Ghana. BMC Pregnancy Childbirth. 2020;20(1):239. PubMed | Google Scholar
- Odoki M, Aliero AA, Tibyangye J, Maniga JN, Eilu E, Ntulume I et al. Fluoroquinolone resistant bacterial isolates from the urinary tract among patients attending hospitals in Bushenyi District, Uganda. PAMJ. 2020;36:1-12. PubMed | Google Scholar
- Bediako-Bowan AAA, Kurtzhals JAL, Mølbak K, Labi A-K, Owusu E, Newman MJ. High rates of multi-drug resistant gram-negative organisms associated with surgical site infections in a teaching hospital in Ghana. BMC Infect Dis. 2020;20(1):890. PubMed | Google Scholar
- Stapleton AE, Wagenlehner FME, Mulgirigama A, Twynholm M. Escherichia coli resistance to fluoroquinolones in community-acquired uncomplicated urinary tract infection in women: a systematic review. Antimicrob Agents Chemother. 2020;64(10):1-20. PubMed | Google Scholar
- Bidell MR, Palchak M, Mohr J, Lodise TP. Fluoroquinolone and third-generation-cephalosporin resistance among hospitalized patients with urinary tract infections due to Escherichia coli: do rates vary by hospital characteristics and geographic region? Antimicrob Agents Chemother. 2016;60(5):3170-3173. PubMed | Google Scholar
- Quansah E, Amoah Barnie P, Omane Acheampong D, Obiri-Yeboah D, Odarkor Mills R, Asmah E et al. Geographical distribution of ß-lactam resistance among Klebsiella spp. from selected health facilities in Ghana. TropicalMed. 2019;4(3):117. PubMed | Google Scholar
- Newman MJ, Frimpong E, Donkor E, Opintan JA, Asamoah-Adu A. Resistance to antimicrobial drugs in Ghana. IDR. 2011;4:215-220. PubMed | Google Scholar
- Vikesland P, Garner E, Gupta S, Kang S, Maile-Moskowitz A, Zhu N. Differential drivers of antimicrobial resistance across the World. Acc Chem Res. 2019;52(4):916-924. PubMed | Google Scholar
- Shariff VAAR, Shenoy MS, Yadav T, Radhakrishna M. The antibiotic susceptibility patterns of uropathogenic Escherichia coli, with special reference to the fluoroquinolones. JCDR. 2013;7(6):1027-1030. PubMed | Google Scholar
- Cardoso T, Ribeiro O, Aragăo IC, Costa-Pereira A, Sarmento AE. Additional risk factors for infection by multidrug-resistant pathogens in healthcare-associated infection: a large cohort study. BMC Infect Dis. 2012;12(1):375. PubMed | Google Scholar
- Mensah-Attipoe I, Opintan JA, Newman MJ, Ashong PP. Molecular characterization of ciprofloxacin resistant Escherichia coli from Ghana. JAMB. 2020;20(10):22-33. PubMed | Google Scholar
- Ogbolu D, Alli A, Anorue M, Daini O, Oluwadun A. Distribution of plasmid-mediated quinolone resistance in Gram-negative bacteria from a tertiary hospital in Nigeria. Indian J Pathol Microbiol. 2016;59(3):322. PubMed | Google Scholar
- Goudarzi M, Azad M, Seyedjavadi SS. Prevalence of plasmid-mediated quinolone resistance determinants and OqxAB efflux pumps among extended-spectrum ß-lactamase producing Klebsiella pneumoniae isolated from patients with nosocomial urinary tract infection in tehran, Iran. Scientifica (Cairo) . 2015;2015:518167. PubMed | Google Scholar