Prevalence of Cryptosporidium parvum, Giardia intestinalis and molecular characterization of group A rotavirus associated with diarrhea in children below five years old in Gaborone, Botswana
Lineage Kurenzvi, Teresa Kibirige Sebunya, Tidimalo Coetzee, Giacomo Maria Paganotti, Mathias Vondee Teye
Corresponding author: Tidimalo Coetzee, Department of Biological Sciences, Faculty of Science, University of Botswana, Gaborone, Botswana 
Received: 07 Aug 2020 - Accepted: 26 Sep 2020 - Published: 14 Oct 2020
Domain: Epidemiology,Microbiology,Parasitology
Keywords: Diarrhea, cryptosporidium, giardia, group A rotavirus, prevalence, genotype
©Lineage Kurenzvi et al. Pan African Medical Journal (ISSN: 1937-8688). This is an Open Access article distributed under the terms of the Creative Commons Attribution International 4.0 License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article: Lineage Kurenzvi et al. Prevalence of Cryptosporidium parvum, Giardia intestinalis and molecular characterization of group A rotavirus associated with diarrhea in children below five years old in Gaborone, Botswana. Pan African Medical Journal. 2020;37:159. [doi: 10.11604/pamj.2020.37.159.25392]
Available online at: https://www.panafrican-med-journal.com//content/article/37/159/full
Research 
Prevalence of Cryptosporidium parvum, Giardia intestinalis and molecular characterization of group A rotavirus associated with diarrhea in children below five years old in Gaborone, Botswana
Prevalence of Cryptosporidium parvum, Giardia intestinalis and molecular characterization of group A rotavirus associated with diarrhea in children below five years old in Gaborone, Botswana
Lineage Kurenzvi1, Teresa Kibirige Sebunya1, Tidimalo Coetzee1,&, Giacomo Maria Paganotti2,3,4, Mathias Vondee Teye1
&Corresponding author
Introduction: Cryptosporidium, Giardia and rotaviruses are amongst the leading causes of acute gastroenteritis in children ≤5 years worldwide. The purpose of this study was to determine the occurrence of Cryptosporidium parvum, Giardia intestinalis and molecular characteristics of rotaviruses after Rotarix® introduction in Botswana.
Methods: in this case study, 200 diarrheic stool specimens and 100 control samples from children under five years old were collected between March and November, 2017. Samples were analyzed by modified Ziehl Neelsen staining technique for cryptosporidium, wet mount procedure for Giardia and negative samples were confirmed by immunochromatographic assay. Specimens were analyzed for rotavirus by ELISA, PAGE, RT-PCR, sequencing of VP7 and VP4 antigen followed by phylogenetic analysis.
Results: prevalence rates of 20.5%, 16.5% and 11.0% in diarrhea cases were observed for Cryptosporidium parvum, Giardia intestinalis and rotavirus, respectively. Four percent of diarrheic specimens had multiple infections. The predominant rotavirus genotype was GIP[8] (7/15) followed by G2P[4] (2/15) and G3P[8] (1/15). Twenty percent of specimens were non-typeable. One mixed strain, G1+G2P[4,8] (2/15), was detected. Phylogenetic analysis of VP4 and VP7 sequences clustered Botswana rotavirus strains within G1 lineages 1 and 2, G3 lineage 1, P[8] lineage 3 and P[4] lineage 5 together with Southern African strains.
Conclusion: this study provides important information on occurrence and demographic risk groups for Cryptosporidium parvum, Giardia intestinalis and rotavirus in young children as well as genetic diversity of rotaviruses after vaccine introduction in Botswana. Constant monitoring of circulating rotavirus strains is essential in assessing effectiveness of current vaccines in Botswana.
Diarrhea is a common disease in developing countries especially in regions where there is poor sanitation and hygiene [1]. In developed countries, diarrhea is rarely fatal except in people at extremes of age and immunocompromised individuals. Globally, most under-fives suffer 3 to 4 episodes of diarrhea in a year [2]. Prolonged and recurrent diarrhea leads to malnutrition and poor growth in children below the age of five years [3]. The introduction of water and sanitation hygiene (WASH) programs, Oral Rehydration Therapy (ORT), vaccinations and adequate nutrition measures gradually reduced mortality and morbidity due to diarrhea in children [4]. However, there are rising concerns about potential increase of diarrheal deaths in Botswana and other nations due to increasing temperatures that pose negative effects on surface water resource [5,6].
Cryptosporidium and Giardia are the most common protozoan pathogens that cause gastroenteritis worldwide. Both parasites are considered emerging opportunistic pathogens responsible for diarrheal morbidity globally [7]. By 2016, Cryptosporidium was the fifth leading cause of diarrheal mortality, responsible for approximately 44.8 million episodes of diarrhea and 48,300 annual deaths in children below the age of five [8]. Prevalence rates of Cryptosporidium ranges from 2 to 60% in Botswana, affecting children ≤24 months of age more than any other age groups [9,10]. This highlights the need to revisit and evaluate the burden of cryptosporidiosis in Botswana.
In 2010, the World Health Organization estimated that Giardia was responsible for 28.2 million cases of diarrhea globally [11]. Giardia outbreaks were also reported in 37% of global waterborne transmission of protozoan parasites between 2011 and 2016 [12]. The occurrence of Giardia ranges from <1% to 72% in Africa [13] and 1 to 10% in Botswana [14,15]. Although prevalence rates of Giardia infections are lower in Botswana, some studies reveal that chronic giardiasis modulates symptoms of rotavirus infection and co-infection in giardiasis patients intensifies severity of diarrhea [16]. Association between Giardia and rotavirus infections has not been well investigated in Botswana, yet higher case fatality ratios have been previously reported in diarrheal children [17].
The Global Burden of Diseases (GBD) reported rotavirus as the leading cause of all diarrhea deaths in 2016, leading to approximately 128,515 deaths in children under five years old [18]. In 2009, two live attenuated oral rotavirus vaccines, RotaTeq™ and Rotarix®, were recommended by World Health Organization in national immunization programmes [19]. The introduction of Rotarix®, increased protection against rotavirus associated illnesses among children in Botswana, but changing diversity of circulating strains had been evident in most studies conducted in the post vaccination era [10,20]. Genetic reassortments of rotavirus strains are possible under vaccine pressure [21] and continuous rotavirus surveillance studies are important in evaluating effectiveness of current vaccines on rotavirus genotypes circulating in Botswana.
In this study, we aimed at determining the prevalence of Giardia intestinalis, Cryptosporidium parvum and rotavirus in children below the age of five years in Gaborone, Botswana. This study was also established to investigate the distribution pattern of group A rotavirus genotypes circulating in young children up to five years of age who were presented with diarrhea in Gaborone.
Study design, study site and study population: this case study was conducted from March to November 2017 in Gaborone, the largest and capital city of Botswana. Diarrheal participants were children below the age of five years whose stool samples were collected at Princess Marina Hospital, Bokamoso Private Hospital and Diagnofirm medical microbiological laboratories. Non-diarrheic participants were children below the age of five from the pediatric ward of Princess Marina Hospital and Gaborone Child Welfare clinics.
Ethical clearance: this study was approved by the Botswana Ministry of Health in Gaborone (ref. no: HPDME 13/18/1), the Institutional Review Board of University of Botswana (ref. no: UBR/RES/IRB/GRAD/297) and Princess Marina Hospital Research and Ethics Committee (ref. no: PMH5/79 [302-1-2017]). Permission was also obtained from all the authorities of wards and clinics. Parental consent was requested from parents/guardians of participants where applicable.
Specimen collection: two hundred stool samples of children presented with diarrhea were collected from Princess Marina Hospital, Bokamoso Private Hospital and Diagnofirm medical microbiological laboratories. For controls, 100 stool specimens from non-diarrheic children were collected from parents/guardians of children from Princess Marina Hospital pediatric ward and Gaborone Child Welfare clinics. Samples were collected in sterile containers, placed in an ice box before transportation to the virology laboratory of University of Botswana. Fecal specimens were divided into two aliquots before storage. Specimens intended to be analyzed for parasites were examined microscopically and stored at -20°C without any preservative. A 10% suspension was made on stool specimens intended to be analyzed for rotavirus. Suspensions were stored at 4°C until the time of analysis.
Detection of Cryptosporidium and Giardia cysts: Giardia cysts were detected microscopically by the wet mount procedure [22]. The modified Ziehl Neelsen technique was used to stain Cryptosporidium oocysts in fecal specimens [23]. All smears were viewed under the light microscope at 10x objective followed by 40x objective. Confirmation of absence of Cryptosporidium parvum and Giardia intestinalis in stool samples that tested negative was done by using a one-step Crypto+Giardia combo card (CerTest Biotec S.L., Zaragoza, Spain).
Laboratory investigations for rotavirus
Polyacrylamide gel electrophoresis: ELISA kits for human rotavirus group A (IVD Research Inc., Carlsbad, CA, USA) were used to detect the presence of rotavirus antigen, following manufacturer´s instructions. Viral RNA was extracted from 16 strongly ELISA positive samples using the phenol/chloroform method. Migration patterns of the rotavirus segmented genome was then detected by polyacrylamide gel electrophoresis (PAGE) in 10% acrylamide slab gels with a 4% stacking gel. A discontinuous buffer system without sodium dodecyl-sulphate was used [24].
Reverse transcriptase-polymerase chain reaction: viral RNA intended to be used for reverse transcriptase-polymerase chain reaction (RT-PCR) was extracted from fecal suspensions using the ZR viral RNA kit (Zymo Research, Orange, CA, USA) following manufacturer´s procedures. The protoscript II cDNA synthesis kit (New England Biolabs, USA) was used for cDNA synthesis according to the manufacturer´s instructions. The cDNA generated from reverse transcription was used as a template for PCR. Specific primer pairs con2/con3 were used for consensus PCR for VP4. Reactions were carried out in a thermal cycler with the following conditions: 2 minutes at 94°C; 35 cycles of 1 minute at 94°C, 1 minute at 50°C, 1 minute at 72°C; a final extended step of 7 minutes at 72°C, then held at 15°C. VP4 genotyping was done by using the forward primer con3 and a cocktail of primers specific to human rotavirus P-genotype P[4], P[6], P[8], P[9] and P[10]. The solution was denatured at 94°C for 4 minutes followed by 30 cycles of PCR at 94°C for 1 minute, 45°C for 2 minutes and 72°C for 1 minute. Reactions were extended for 7 minutes at 72°C, then held at 15°C.
Beg9/End9 primer pair was used for the outer PCR for VP7. Reactions were cycled under the following conditions: 2 minutes at 94°C; 35 cycles of 1 minute at 94°C, 1 minute at 52°C, 1 minute at 72°C; with a final extension of 7 minutes at 72°C, then held at 15°C. Multiplex PCR for VP7 was done using the reverse primer End9 with a cocktail of primers specific to G1, G2, G3, G4, G8 and G9. PCR reactions were performed in a thermal cycler with the following conditions: 2 minutes at 94°C; then 35 cycles of 1 minute at 94°C, 1 minute at 50°C, 1 minute at 72°C; with a final extended step of 7 minutes at 72°C, held at 15°C. All VP4 and VP7 amplicons were visualized by electrophoresis (alongside a 100bp DNA ladder) on a 1.5% agarose gel stained with ethidium bromide then photographed under ultraviolet light.
Nucleotide sequencing of RT-PCR amplicons and phylogenetic analysis: amplicons from 10 specimens were selected for sequence analysis. Sequencing was done using the same primers for RT-PCR. PCR products were purified by the Exonuclease and Shrimp Alkaline Phosphatase (ExoSAP) procedure according to manufacturer´s protocol. Fragments were sequenced using the nimagen, BrilliantDye™ terminator cycle sequencing kit V3.1, according to manufacturer´s instructions. Labelled products were then cleaned with the ZR-96 DNA sequencing clean-up kit (Zymo Research, Orange, CA, USA) and later injected on the Applied Biosystems ABI 3500XL genetic analyser with a 50cm array, using POP7.
Data analysis
Statistical analysis: to summarize data generated from serological tests and electrophoresis, descriptive statistics (frequencies and percentages) were calculated for all categorical variables using Microsoft Excel Software, Version 2013 (Microsoft Corporation, USA). The Chi-square test was used to compare categorical variable proportions and statistical significance. A P-value of less than 0.05 was considered to be statistically significant.
Phylogenetic analysis: after sequencing, the chromatograms of sequences were assembled and visually analyzed using BioEdit software version 7.2.5 [25]. Identification of genotypes of the sequences was achieved by comparison with reference sequences available in the NCBI GenBank database using BLAST [26]. Multiple sequence alignments was carried out using Clustal W [27] incorporated in BioEdit software version 7.2.5 [25]. Phylogenetic analysis was done in MEGA software version 6.06 [28] using a distance-based neighbor-joining of the Kimura 2-parameter nucleotide substitution model [29]. The robustness of each tree branch was tested by performing 1,000 bootstrap replicates.
Nucleotide sequence accession numbers: the nucleotide sequences of group A rotaviruses of this study were deposited in GenBank and assigned accession numbers MT364283 to MT364302 (Table 1).
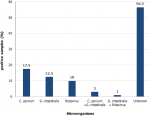
Epidemiology of Cryptosporidium parvum, Giardia intestinalis and rotavirus: the most common pathogens in this study were parasites (Figure 1). Cryptosporidium parvum was detected in 20.5% (41/200) of all the diarrheic samples, and 61% (25/41) of positive samples came from male children. Most Cryptosporidium parvum cases were from children ≤36 months of age (32/41, 78%, P=0.100). The peak month of cryptosporidium infections was October and cases dropped during the winter season. Giardia intestinalis was found in 16.5% (33/200) of children suffering from diarrhea. No significant association was found between prevalence and gender, although it affected more females than males (18/33, 54.5%, P=0.895). More cases of Giardia intestinalis were from children aged ≤36 months (27/33, 81.8%, P=0.449). Slight fluctuations in occurrence of Giardia intestinalis were seen frequently throughout the entire study period.
Rotavirus was the least detected pathogen (11.0%) in all 200 samples of children with diarrhea. Fifteen (15) samples (68.2%, P=0.93) were from children aged ≤24 months old. Rotavirus affected male (11/22, 50%) and females (11/22, 50%) equally. Infections were at peak in April and in July. Three percent (3%) of diarrheic samples yielded both Giardia intestinalis and Cryptosporidium parvum, while 1% had both Giardia intestinalis and rotavirus.
Rotavirus electrophoretypes and genotypes: most of the isolates revealed the 4-2-3-2 gene migration pattern of human group A rotavirus. Only two major patterns were identified from diarrheic samples, long (9/16, 56.25%) and short (4/16, 25%) electrophoretypes. Two samples (12.5%) had more than 11 gene segments and no clear migration was found in 6.25% (1/15) of diarrheic samples. Only 3 G genotypes, G1 (9/15, 60%), G2 (4/15, 26.7%) and G3 (1/15, 6.7%) were observed. The most predominant genotype was GIP[8] (6/15, 40%) followed by G2P[4] (2/15, 13.3%) and G3P[8] (1/15, 6.7%). Uncommon rotavirus genotype G1P[6] was detected in approximately 7% (1/15) by RT-PCR. Two cases (13.3%) were found to have an uncommon combination G1+G2P [4,8] and 20% of the specimens tested by PCR were untypeable.
Phylogenetic analysis of VP4 and VP7 genes: sequence alignment and phylogenetic analysis of the VP7 genes of the 10 rotavirus strains that were successfully sequenced showed 96-99% nucleotide sequence identity after being compared to corresponding G1 and G3 strains internationally. The VP4 gene sequences of rotavirus strains from this study showed nucleotide sequences with 96-97% identity with regional and international P[4] and P[8] strains. One sample that was thought to be G1P[6] by RT-PCR was confirmed to be G1P[8] after sequencing. Sequencing of the G2 strains detected by RT-PCR was not successful, therefore no sequence data was obtained for phylogenetic analysis.
The G1 strains of Botswana clustered G1 sequences into 2 lineages (Figure 2). Five of the G1 strains (RVA/human-wt/BWA/O14/2017/G1, RVA/human-wt/063/2017/G1, RVA/human-wt/BWA/141/2017/G1, RVA/human-wt/BWA/145/2017/G1 and RVA/human-wt/BWA/161/2017/G1) clustered in lineage 1 with strains from South Africa (KJ753805, KJ752278, KJ753123 and KP753022), Malawi (MG181474, MG181331 and MG181507), Mozambique (KP222814, KP222809), Thailand (DQ512974, DQ512981), India (JN192064) and Belgium (JN849122). The other four G1 sequences (Figure 2) from this current study: RVA/human-wt/BWA/048a/2107/G1, RVA/human-wt/BWA075a/2017/G1, RVA/human-wt/BWA/163/2017/G1 and RVA/human-wt/BWA/165/2017/G1 fell into lineage 2 and exhibited 97 to 99% identity among themselves and clustered with strains discovered earlier in India (KX638546), South Africa (KJ751751) and China (DQ873669). The G3 sequence of this study (RVA/human-wt/BWA/149/2017/G3) was assigned to G3 lineage 1 and was closely related to G3 strains previously identified in South Africa (KP752598, KJ753186 and KJ753440) and China (DQ873669) (Figure 2). A distant relationship was found between all VP7 strains with both Rotarix (G1 lineage 2) and RotaTeq (G3 lineage 2).
Botswana VP4 sequences (Figure 3) were assigned to P[8] genotype lineage 3 and P[4] lineage 5 and were all distantly related to both Rotarix (P[8] lineage 2) and RotaTeq (P[8] lineage 1). All the observed P[8] sequences in this study shared 100% nucleotide sequence identity with each other and clustered in P[8] lineage 3 with sequences from South Africa (KJ753803, KJ753121), Malawi (MG181483), Gambia (KJ752287), Senegal (KJ751560) and Belgium (HQ392119, JN849147). Isolated P[4] strains (RVA/human-wt/BWA/048b/2017/P4 and RVA/human-wt/BWA/075b/2017/P4) clustered with other sequences from Malawi (MG181912 and MG181835), Mauritius (KP752663) and Belgium (KR705171).
This present study determines the prevalence of Cryptosporidium parvum, Giardia intestinalis and rotavirus among children below the age of five years in Gaborone. Prevalence of 20.5% for Cryptosporidium parvum and 16.5% for Giardia intestinalis observed in this study were higher than those reported in similar studies previously conducted in Botswana [10,14,15]. A possible explanation of this discrepancy can be variations in research set ups, sample population, environmental condition of study locations, sample size as well as methods used for detection. Furthermore, climate change recently increased water scarcity in cities like Gaborone, promoting the use of contaminated household water storage and increase exposure to parasitic infections [30].
Most Cryptosporidium parvum and Giardia intestinalis infections occurred primarily in children ≤36 months of age, and this correlates with worldwide reports that cryptosporidiosis and giardiasis affects children in lower age groups more frequently than any other age group [8]. High prevalence of Giardia intestinalis and Cryptosporidium parvum in younger children may be as a result of increased exposure to contaminated municipal water and increased contact with contaminated surfaces. Lack of previous exposure to Giardia intestinalis also render young children to be more susceptible to infection [31].
Approximately 60% of male children were significantly affected by cryptosporidiosis. Most researchers associate cryptosporidiosis and giardiasis distribution to social, cultural and behavioral differences between male and female children [32,33]. However, no apparent association was established between gender and giardiasis in this study. Lack of association of prevalence of Giardia intestinalis with gender might have been caused by a smaller sample size used in this current study. Although giardiasis show fluctuations from spring to winter, the peak months of both cryptosporidiosis and giardiasis transmission coincided with the hot rainy season. This finding agrees with prior reports from Chobe district that suggested that cryptosporidiosis and giardiasis coincide with high precipitation periods [5].
In Botswana, rotavirus has been reported as the most prevalent pathogen of diarrhea in studies where both Cryptosporidium species and rotavirus had been investigated before rotavirus vaccine introduction [34]. Lower prevalence of rotavirus than Cryptosporidium observed in this study can be as a result of high efficiency of the monovalent rotavirus vaccine Rotarix® currently used in Botswana [20]. Similar trends were reported in other African countries that engaged Rotarix in their immunization programmes [35,36]. A higher proportion of rotavirus in children ≤24 months old observed in this current study is consistent with some previous studies in Botswana [20,34] and other nations like South Rajasthan and China [37,38]. Peak prevalence of rotavirus infections in colder months agree with previous findings in local studies [39-41] although some researchers deduced that there is no unifying explanation for the global varying seasonality of rotavirus [42].
Only 3 G genotypes (G1, G2 and G3) and 2 P genotypes (P[4] and P[8]) were observed in this study polyacrylamide gel electrophoresis confirmed that all the G1 were associated with long electrophoretypes whilst all G2 and G3 showed short electrophoretic patterns. These migration pattern correlate with findings from other researchers in Botswana [40,41], Eastern and Southern African countries [43]. The most predominant genotype of rotavirus in this study was G1P[8], detected in 40% of all sequenced samples. In Botswana G1P[8] was one of the major isolated strains before rotavirus vaccination [41]. G1P[8] gradually decreased in Botswana after Rotarix® introduction, but later reappeared the following year [44]. Global epidemiological studies on rotavirus also identified G1P[8] as one of the most prevalent strain even after vaccine introduction [45]. Phylogenetic analysis of G1 sequences assigned five of Botswana G1P[8] strains into lineage 1 and were closely related to some G1P[8] strains discovered in South Africa, Malawi and Mozambique. This observation indicates that these G1P[8] strains might be direct descendants of previous strains from neighboring countries rather than emergence from point mutations. The other four G1 sequences formed a separate cluster in lineage 2 together with G1P[8] strains previously observed in India. This observation indicates that these might have been introduced in Botswana by continental migration, genetic reassortment or mutation.
In this study, G2P[4] was the second leading cause of rotavirus associated diarrhea, although it was the leading cause of rotavirus acquired illnesses in a post vaccination surveillance study in Botswana [20]. Circulating rotavirus genotypes may vary on a yearly basis and their occurrence may also be affected by natural cyclic patterns as well as seasonal variations [46,47], highlighting the importance of surveillance studies of rotavirus diseases. Phylogenetic analysis of the P[4] sequences clustered the G2P[4] strains of the current study in P[4] lineage 2 with other strains from Malawi, Mauritius and Belgium, indicating the derivation of those strains from a common origin. However, phylogenetic analysis of G2 sequences of this study was impossible. This might have been caused by poor DNA quality and low viral titers. G3 strain for human rotavirus was first detected in Botswana in the early 2000s and in most cases was associated with P[6] and other untypeable P genotypes [40,41]. G3 cases drastically declined in the post vaccination era [20] and was the least detected in this present study. Similar observations were reported in studies conducted in Eastern and Southern African countries after Rotarix vaccine introduction [43]. This suggests that Rotarix® may have high efficacy against G3P[8] as well as cross-protection against serotypes not included in the vaccine´s composition. The single G3P[8] of this current study was closely related to G3P[8] strains circulating in South Africa and China indicating that they might have descended from a common ancestor.
In many African countries, rotavirus strains of G types G1, G2, G8, G9 and P types P[4], P[6], P[8], P[9] had been isolated from drinking water, which is one of the major environmental factors that promote mixed infections in developing countries [48,49]. Phylogenetic analysis of samples with mixed infections of this study revealed that G1+G2 P[4,8] was as a result of infection by both G1P[8] and G2P[4] strains closely related to previously identified strains from South Africa and Malawi, respectively. However, complete genome sequencing of the strains would be a necessity to determine the potential of any natural reassortment.
Cryptosporidium parvum, Giardia intestinalis and rotavirus are most important pathogens of diarrheal diseases in young children. Most Cryptosporidium parvum and Giardia intestinalis transmissions coincided with the hot, rainy season of the year while rotavirus infections were at peak in winter. Further research is required to understand seasonal peaks of Cryptosporidium parvum, Giardia intestinalis and rotavirus to establish more intervention measures that disrupt transmission in young children. Rotavirus strains circulating in Botswana are similar to those frequently occurring worldwide. Phylogenetic analyses of the isolated rotavirus strains of Botswana indicate that they are distantly related to Rotarix, but there might be transmission between Botswana and some southern African countries, especially South Africa and Malawi. To obtain more understanding of the epidemiology of rotavirus strains in the region and to assess the effectiveness of vaccines strains currently being used in the country, more and continuous surveillance studies of circulating rotavirus genomes need to be done. Whole genome sequencing of circulating rotavirus strains would be essential to determine the extent of genetic variation and the relatedness of Botswana rotavirus strains with those described worldwide.
What is known about this topic
- Cryptosporidium, Giardia and rotaviruses mostly affects children younger than 2 years in some parts of Botswana;
- The distribution of rotavirus genotypes circulating in young children changed after vaccine implementation in Botswana.
What this study adds
- This study confirmed that Cryptosporidium, Giardia and rotavirus are more prevalent in the early stages of life than the later;
- This study provides information on the sequences of circulating rotavirus strains in young children in Gaborone in 2017;
- This present study confirmed that most rotavirus strains circulating in children below 5 years old in Gaborone are related to strains previously identified in other Southern African countries.
The authors declare no competing interests.
LK and TKS conceived and designed the study; TKS and TTC supervised the project; LK collected the samples and together with GMP performed laboratory analyses; LK and MVT performed data analysis; LK drafted the manuscript; TKS, TTC, GMP and MVT proofread and revised the manuscript. All authors attest that they meet the ICMJE criteria for authorship. All the authors have read and agreed to the final manuscript.
The authors would like to acknowledge all the microbiology laboratory staff at Princess Marina Hospital, Bokamoso Private Hospital, Diagnofirm and Gaborone Child Welfare Clinics for all their assistance during sampling time. We thank Inqaba Biotechnological Laboratory staff for their contributions on this study.
Table 1: VP7 and VP4 sequences obtained in this study and their GenBank accession numbers
Figure 1: prevalence of Cryptosporidium parvum, Giardia intestinalis and rotaviruses in children under five years old in Gaborone
Figure 2: phylogenetic analysis of VP7 nucleotide sequences of rotaviruses in Botswana and international strains based on the neighbor-joining method; the 2017 strains from Botswana are indicated by red diamonds; vaccine strains are indicated by circles; bootstrap test (1000 replicates) values are shown on the branches
Figure 3: phylogenetic analysis of VP4 nucleotide sequences of rotaviruses in Botswana and international strains based on the neighbor-joining method; the 2017 strains from Botswana are indicated by red diamonds; vaccine strains are indicated by circles; bootstrap test (1000 replicates) values are shown on the branches
- Levine GA, Walson JL, Atlas HE, Lamberti LM, Pavlinac PB. Defining pediatric diarrhea in low-resource settings. Journal of the Pediatric Infectious Diseases Society. 2017;6(3):289-293. PubMed | Google Scholar
- Walker CLF, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA et al. Global burden of childhood pneumonia and diarrhoea. The Lancet. 2013;381(9875):1405-1416. PubMed | Google Scholar
- Pinkerton R, Oriá RB, Lima AA, Rogawski ET, Oriá MO, Patrick PD et al. Early childhood diarrhea predicts cognitive delays in later childhood independently of malnutrition. The American Journal of Tropical Medicine and Hygiene. 2016;95(5):1004-1010. PubMed | Google Scholar
- Darvesh N, Das JK, Vaivada T, Gaffey MF, Rasanathan K, Bhutta ZA et al. Water, sanitation and hygiene interventions for acute childhood diarrhea: a systematic review to provide estimates for the Lives Saved Tool. BMC Public Health. 2017;17(Suppl 4):776. PubMed | Google Scholar
- Alexander KA, Carzolio M, Goodin D, Vance E. Climate change is likely to worsen the public health threat of diarrheal disease in Botswana. International Journal of Environmental Research and Public Health. 2013;10(4):1202-1230. PubMed | Google Scholar
- Carlton EJ, Woster AP, DeWitt P, Goldstein RS, Levy K. A systematic review and meta-analysis of ambient temperature and diarrhoeal diseases. International Journal of Epidemiology. 2016;45(1):117-130. PubMed | Google Scholar
- Troeger C, Blacker BF, Khalil IA, Rao PC, Cao S, Zimsen SR et al. Estimates of the global, regional and national morbidity, mortality and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. The Lancet Infectious Diseases. 2018;18(11):1211-1228. PubMed | Google Scholar
- Khalil IA, Troeger C, Rao PC, Blacker BF, Brown A, Brewer TG et al. Morbidity, mortality and long-term consequences associated with diarrhoea from cryptosporidium infection in children younger than 5 years: a meta-analyses study. The Lancet Global Health. 2018;6(7):758-768. PubMed | Google Scholar
- Creek TL, Kim A, Lu L, Bowen A, Masunge J, Arvelo W et al. Hospitalization and mortality among primarily nonbreastfed children during a large outbreak of diarrhea and malnutrition in Botswana, 2006. J Acquir Immune Defic Syndr. 2010;53(1):14-19. PubMed | Google Scholar
- Goldfarb DM, Steenhoff AP, Pernica JM, Chong S, Luinstra K, Mokomane M et al. Evaluation of anatomically designed flocked rectal swabs for molecular detection of enteric pathogens in children admitted to hospital with severe gastroenteritis in Botswana. Journal of Clinical Microbiology. 2014;52(11):3922-3927. PubMed | Google Scholar
- World Health Organization. World health statistics 2015. World Health Organization. 2015. Google Scholar
- Efstratiou A, Ongerth JE, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks-an update 2011-2016. Water Research. 2017;114:14-22. PubMed | Google Scholar
- Squire SA, Ryan U. Cryptosporidium and giardia in Africa: current and future challenges. Parasites and Vectors. 2017;10(1):195. PubMed | Google Scholar
- Alexander KA, Herbein J, Zajac A. The occurrence of cryptosporidium and giardia infections among patients reporting diarrheal disease in Chobe district, Botswana. Advances in Infectious Diseases. 2012;2(4):143-147. Google Scholar
- Rowe JS, Shah SS, Motlhagodi S, Bafana M, Tawanana E, Truong HT et al. An epidemiologic review of enteropathogens in Gaborone, Botswana: shifting patterns of resistance in an HIV endemic region. PloS One. 2010;5(6):e10924. PubMed | Google Scholar
- Bhavnani D, Goldstick JE, Cevallos W, Trueba G, Eisenberg JN. Synergistic effects between rotavirus and coinfecting pathogens on diarrheal disease: evidence from a community-based study in northwestern Ecuador. American Journal of Epidemiology. 2012;176(5):387-395. PubMed | Google Scholar
- Welch H, Steenhoff AP, Chakalisa UA, Arscott-Mills T, Mazhani L, Mokomane M et al. Hospital-based surveillance for rotavirus gastroenteritis using molecular testing and immunoassay during the 2011 season in Botswana. The Pediatric Infectious Disease Journal. 2013;32(5):570-572. PubMed | Google Scholar
- Troeger C, Khalil IA, Rao PC, Cao S, Blacker BF, Ahmed T et al. Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. Journal of American Medical Association Pediatrics. 2018;172(10):958-965. PubMed | Google Scholar
- Tate JE, Patel MM, Steele AD, Gentsch JR, Payne DC, Cortese MM et al. Global impact of rotavirus vaccines. Expert Review of Vaccines. 2010;9(4):395-407. PubMed | Google Scholar
- Gastañaduy PA, Steenhoff AP, Mokomane M, Esona MD, Bowen MD, Jibril H et al. Effectiveness of monovalent rotavirus vaccine after programmatic implementation in Botswana: a multisite prospective case-control study. Clinical Infectious Diseases. 2016;62(suppl_2):S161-S167. PubMed | Google Scholar
- Kirkwood CD. Genetic and antigenic diversity of human rotaviruses: potential impact on vaccination programs. Journal of Infectious Diseases. 2010;202(suppl_1):S43-S48. PubMed | Google Scholar
- Baig MF, Kharal SA, Qadeer SA, Badvi JA. A comparative study of different methods used in the detection of giardia lamblia on fecal specimens of children. Annals of Tropical Medicine and Public Health. 2012;5(3):163. Google Scholar
- Fayer R, Morgan U, Upton SJ. Epidemiology of cryptosporidium: transmission, detection and identification. International Journal for Parasitology. 2000;30(12-13):1305-1322. PubMed | Google Scholar
- World Health Organization. Manual of rotavirus detection and characterization methods, 2009. Geneva: World Health Organization. 2016;8(17):6-99.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41(41):95-98. Google Scholar
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403-410. PubMed | Google Scholar
- Thompson JD, Higgins GD, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673-80. PubMed | Google Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6. Molecular Biology and Evolution. 2013;30(12):2725-2729. PubMed | Google Scholar
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16(2):111-120. PubMed | Google Scholar
- Kadibadiba T, Roberts L, Duncan R. Living in a city without water: a social practice theory analysis of resource disruption in Gaborone, Botswana. Global environmental change. 2018;53:273-285. Google Scholar
- Painter JE, Gargano JW, Collier SA, Yoder JS, Centers for Disease Control and Prevention. Giardiasis surveillance-United States, 2011-2012. Morbidity and Mortality Weekly Reports: Surveillance Summaries. 2015;64(3):15-25. PubMed | Google Scholar
- Al Saqur IM, Al-Warid HS, Albahadely HS. The prevalence of giardia lamblia and entamoeba histolytica/dispar among Iraq provinces. Karbala International Journal of Modern Science. 2017;3(2):93-96. Google Scholar
- Lal A, Fearnley E, Kirk M. The risk of reported cryptosporidiosis in children aged <5 years in Australia is highest in very remote regions. International Journal of Environmental Research and Public Health. 2015;12(9):11815-11828. PubMed | Google Scholar
- Pernica JM, Steenhoff AP, Welch H, Mokomane M, Quaye I, Arscott-Mills T et al. Correlation of clinical outcomes with multiplex molecular testing of stool from children admitted to hospital with gastroenteritis in Botswana. Journal of the Pediatric Infectious Diseases Society. 2016;5(3):312-318. PubMed | Google Scholar
- Madhi SA, Cunliffe NA, Steele D, Witte D, Kirsten M, Louw C et al. Research article (New England Journal of Medicine): effect of human rotavirus vaccine on severe diarrhea in African infants. Malawi Medical Journal. 2016;28(3):108-114. PubMed | Google Scholar
- Tate JE, Ngabo F, Donnen P, Gatera M, Uwimana J, Rugambwa C et al. Effectiveness of pentavalent rotavirus vaccine under conditions of routine use in Rwanda. Clinical Infectious Diseases. 2016;62(suppl_2):S208-S212. PubMed | Google Scholar
- Ameta P, Nayak VH, Goyal SC. Prevalence and seasonal distribution of rotavirus diarrhea in hospitalized children less than 5 year old in South Rajasthan. Int J Biomed Res. 2015;6(3):214-218. Google Scholar
- Wu D, Yen C, Yin ZD, Li YX, Liu N, Liu YM et al. The public health burden of rotavirus disease in children younger than five years and considerations for rotavirus vaccine introduction in China. The Pediatric Infectious Disease Journal. 2016;35(12):e392-e398. PubMed | Google Scholar
- Basu G, Rossouw J, Sebunya TK, Gashe BA, De Beer M, Dewar JB et al. Prevalence of rotavirus, adenovirus and astrovirus infection in young children with gastroenteritis in Gaborone, Botswana. East African Medical Journal. 2003;80(12):652-655. PubMed | Google Scholar
- Kebaabetswe LP, Sebunya TK, Matsheka MI, Ndung'u T. Detection and molecular characterisation of group A rotavirus from children in northern Botswana. East African Medical Journal. 2005;82(4):203-208. PubMed | Google Scholar
- Kasule M, Sebunya TK, Gashe BA, Armah G, Steele AD. Detection and characterization of human rotavirus among children with diarrhoea in Botswana. Tropical Medicine and International Health. 2003;8(12):1137-1142. PubMed | Google Scholar
- Baker JM, Alonso W. Rotavirus vaccination takes seasonal signature of childhood diarrhea back to pre-sanitation era in Brazil. Journal of Infection. 2018;76(1):68-77. PubMed | Google Scholar
- Seheri LM, Magagula NB, Peenze I, Rakau K, Ndadza A, Mwenda JM et al. Rotavirus strain diversity in Eastern and Southern African countries before and after vaccine introduction. Vaccine. 2018;36(47):7222-7230. PubMed | Google Scholar
- Mokomane M, Esona MD, Bowen MD, Tate JE, Stennhoff AP, Lechiile K et al. Diversity of rotavirus strains circulating in Botswana before and after introduction of the monovalent rotavirus vaccine. Vaccine. 2019;37(43):6324-6328. PubMed | Google Scholar
- Luchs A, Timenetsky MDCST. Group A rotavirus gastroenteritis: post-vaccine era, genotypes and zoonotic transmission. Einstein (Sao Paulo). 2016;14(2):278-287. PubMed | Google Scholar
- Matthijnssens J, Nakagomi O, Kirkwood CD, Ciarlet M, Desselberger U, Ranst MV. Group A rotavirus universal mass vaccination: how and to what extent will selective pressure influence prevalence of rotavirus genotypes. Expert Review of Vaccines. 2012;11(11):1347-1354. PubMed | Google Scholar
- Mwenda JM, Tate JE, Parashar UD, Mihigo R, Agócs M, Serhan F et al. African rotavirus surveillance network: a brief overview. The Pediatric Infectious Disease Journal. 2014;33(1):S6-S8. PubMed | Google Scholar
- Chigor V, Okoh A. Quantitative RT-PCR detection of hepatitis A virus, rotaviruses and enteroviruses in the Buffalo River and source water dams in the Eastern Cape Province of South Africa. International Journal of Environmental Research and Public Health. 2012;9(11):4017-4032. PubMed | Google Scholar
- Aminu M, Page NA, Ahmad AA, Umoh JU, Dewar J, Steele AD. Diversity of rotavirus VP7 and VP4 genotypes in Northwestern Nigeria. Journal of Infectious Diseases. 2010;202(suppl_1):S198-S204. PubMed | Google Scholar
Search
This article authors
On Pubmed
On Google Scholar
Citation [Download]
Navigate this article
Similar articles in
Key words
Tables and figures
 Figure 2: phylogenetic analysis of VP7 nucleotide sequences of rotaviruses in Botswana and international strains based on the neighbor-joining method; the 2017 strains from Botswana are indicated by red diamonds; vaccine strains are indicated by circles; bootstrap test (1000 replicates) values are shown on the branches
Figure 2: phylogenetic analysis of VP7 nucleotide sequences of rotaviruses in Botswana and international strains based on the neighbor-joining method; the 2017 strains from Botswana are indicated by red diamonds; vaccine strains are indicated by circles; bootstrap test (1000 replicates) values are shown on the branches
 Figure 3: phylogenetic analysis of VP4 nucleotide sequences of rotaviruses in Botswana and international strains based on the neighbor-joining method; the 2017 strains from Botswana are indicated by red diamonds; vaccine strains are indicated by circles; bootstrap test (1000 replicates) values are shown on the branches
Figure 3: phylogenetic analysis of VP4 nucleotide sequences of rotaviruses in Botswana and international strains based on the neighbor-joining method; the 2017 strains from Botswana are indicated by red diamonds; vaccine strains are indicated by circles; bootstrap test (1000 replicates) values are shown on the branches